Autor : Imperiale BelĂ©n1, 2, Di Giulio Beatriz3, Moyano Roberto D.1, 2, Santangelo Ma de la Paz1, 2, Tártara Silvina4, Alonso Viviana4, Sanjurjo Myriam4, GarcĂa Graciela4, Castellaro Patricia4, Romano Ma Isabel1, 2, Morcillo Nora4
1 Instituto de BiotecnologĂa, Instituto Nacional de TecnologĂa Agropecuaria (INTA/CICV), Buenos Aires 2 National Scientific and Technical Research Council (CONICET), CABA 3 Hospital Petrona V. de Cordero, San Fernando, Buenos Aires 4Hospital Dr. Antonio A. Cetrángolo, Vicente LĂłpez, Buenos Aires, Argentina
Correspondencia : Nora Morcillo nora_morcillo@yahoo.com.ar
Abstract
Introduction: The term non-tuberculous mycobacteria (NTM) includes different ambient species capable of sickening humans and/or animals, even by means of a potential
zoonotic transmission.
Objectives: To determine: The clinical importance of several species within the genus Mycobacterium and the genetic diversity of the M. avium complex (MAC), the in vitro bacterial sensitivity and the success of the specific treatment.
Materials and Methods: Collection of clinical and epidemiologic data and information
about isolates of the 2009-2016 period; molecular identification of the isolates; determination of the in vitro bacterial sensitivity and genetic diversity of the MAC; treatment
evaluation.
Results: 225 mycobacteriosis cases were diagnosed, with a stable prevalence of ≈6%
per year and 22 recovered species: 4 rapidly growing species isolated from 66 patients
and 18 slowly growing species. The MAC was isolated in 95 cases, M. avium hominissuis - 40 cases, M. intracellulare - 51 cases, M. chimaera - 3 cases and M. colombiense - 1
case. We observed a greater probability of getting sick from M. intracellulare in patients
previously treated for tuberculosis (TB). HIV-positive patients had a greater risk of falling ill from M. avium hominissuis. Aminoglycosides, fluoroquinolones and macrolides
were the most active drugs against most NTM. Approximately half of the cases healed.
Conclusions: M. intracellulare, M. aviumhominissuis with great genetic variability and M. abscessus were the most commonly found pathogens. The cases of TB+NTM mixed
disease were an important finding. For treating these patients, it was necessary to add
second line drugs to the therapeutic regimen for TB; and most of them healed.
Key words: Non-tuberculous mycobacteria; Diversity and clinical importance
Introduction
The term non-tuberculous mycobacteria (NTM)
includes several species capable of producing
a broad spectrum of pathogens and playing a
potential zoonotic role in animals and humans.
Some species are strictly pathogenic, whereas
other species may cause opportunistic infections,
like the members of the Mycobacterium avium
complex (MAC). However, during the last decades, there have been more cases of the disease caused
by NTM, or mycobacteriosis, and more conditions
causing immunosuppression, such as HIV infection, and malignant and autoimmune diseases,
whose treatment generate the conditions necessary for an NTM infection to develop to a disease
in infected individuals1,2.
Rapidly growing mycobacteria (RGM) were defined as the species that need less than 7 days to
produce colonies easily observed with the naked eye in solid culture media3. This group includes approximately 56 ambient species. Some of them may
be isolated from humans or animals. M. fortuitum, M. abscessus, M. chelonae, M. smegmatis, M. immunogenum, M. peregrinum, M. houstonense, M.
neworleansense (M. mageritense, M. septicum, M.
mucogenicum [previously M. chelonae-like organisms]), M. wolinskyi and M. goodii were isolated
from patients4-6. M. porcinum, M. farcinogenes and M. senegalense are regarded as pathogens of
veterinary importance, though M. porcinum was
responsible for an outbreak in humans related to
the supply of drinking water7. A few years ago, M. abscessus was subdivided into three genome
species, and more recently, Totrtoli et al divided
it into subspecies of the M. abscessus complex: M.
abscessus subspecies abscessus, M. abscessus subspecies masiliense and M. abscessus subspecies bolletii8-10. For semantic simplicity reasons, some of
the mentioned species are frequently grouped and
referred to as groups or complexes: M. abscessus,
including said species, and M. fortuitum-chelonae,
including the species M. chelonae, M. senegalense,
M. smegmatis and M. fortuitum7, 10, 11.
On the other hand, slowly growing mycobacteria (SGM) are the species that need more than
seven days to show visible growth in solid culture
media2,3. The MAC includes: M. avium subspecies selvaticum, M. avium subspecies hominissuis, M. avium subspecies avium, M. avium subspecies paratuberculosis, M. intracellulare and the
recently added species, M. chimaera and M. colombiense12-14. M. avium subspecies hominissuis
and M. intracellulare are the most common and
important human pathogens, whereas M. paratuberculosis (MAP) is a very important bovine
pathogen that causes Paratuberculosis or Johne disease in animals. This mycobacterium has been
suggested as a pathogenic agent of Chron’s disease
in humans12,13.
Recently disseminated studies about the complete sequencing of a total of 120 genomes of
strains considered as individual species showed
that, in some cases, they could be either previously identified species or other species with a very
close phylogenetic relationship with them. Thus, M. nonchromogenicum and M. kumamotonense have been included in the M. terrae complex; M.
chimaera and M. colombiense have been included
in the MAC; and M. ulcerans and M. pseudozhottsii have been included in the M. marinum complex14,15.
Unlike what happens with the microbiological
diagnosis of tuberculosis (TB), and given the natural habitat of NTM that includes the soil and the
water, the microbiological diagnosis of mycobacteriosis must meet certain requirements established
by the American Thoracic Society (ATS), stating if
it is a new colonization or if the isolated mycobacterium is the true cause for the disease15.
The objectives of this study were: To consider
the frequency and clinical importance of NTM as
causative agents; determine the proportion of cases
caused by SGM and RGM; explore the appearance
of more than one mycobacterium isolated either
simultaneously or not simultaneously in the same
host; observe differences in the sensitivity of different NTM species to antibiotics; evaluate the
success of the specific treatment; and analyze the
genetic diversity of the MAC, the main cause for
mycobacteriosis, within the period between January 2009 and December 2016.
Materials and Methods
During the study period, we gathered the following
epidemiologic, demographic and clinical patient
information: age, gender, district of residence;
disease localization according to the images obtained; HIV infection; comorbidities causing an
immunosuppressed state; previous TB or mycobacterioris treatments; and amount of analyzed
clinical materials with bacteriological results.
Respiratory clinical specimens, such as sputum, bronchial and bronchoalveolar lavages and
endotracheal aspirate, and also extrapulmonary
specimens, such as pleural, spinal and abdominal
fluids, stools, urine, tissues, blood and bone marrow were collected.
All the materials, except for the blood and
bone marrow that were incubated with the MycoF-Lytic BACTEC 9050 system (BD Argentina)
were processed for direct microscopic observation by Ziehl-Neelsen staining and cultured in
solid media (Löwenstein-Jensen and Stonebrink)
and in the automated BACTEC MGIT 960TM 16,17
system.
Antimicrobial sensitivity was determined in
vitro by means of a colorimetric micro-method
(resazurin micro assay, REMA) that uses resazurin
as a vital dye and indicates bacterial growth or the
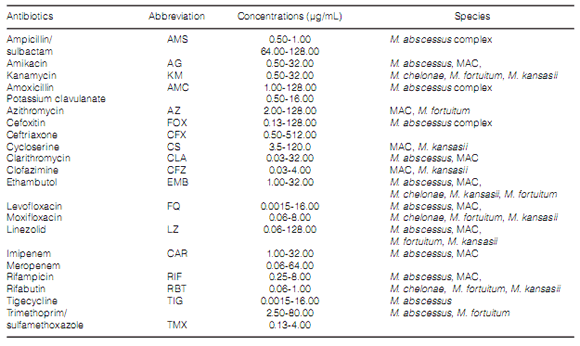
absence of it18. Table 1 shows which drugs were
included in the antibiogram.
The identification of the different found species was carried out through the GenoType CMTM
system19. When identification results were incomplete, we sequenced the rRNA gene in order to
correctly identify the isolates20, 21.
The differentiation of the MAC members was
made through the polymerase chain reaction of
insertion sequence 1311 (PCR IS1311) (21). PCR
IS901 was used to specifically differentiate subspecies M. avium subsp. avium (MAA) from M. avium subsp. Hominissuis22.
The genetic diversity of the MAC was explored
by studying the polymorphism present at 8 loci
with MIRU-VNTR structure (mycobacterial interspersed repetitive units-variable number of
tandem repeats), through the technique previously described by Thibault et al. 2007 (23). The
allelic diversity (D) of every locus and the global
discriminatory power of the MIRU-VNTR complete
scheme (HGDI, Hunter Gaston Discriminatory
Index) were determined with the online software
edited by the Universidad del País Vasco23-25.
http://insilico.ehu.es/mini_tools/discriminatory_power/ through the following formula:
Where N is the total number of isolates of every
type of scheme; s is the total number of different
subtypes discriminated by the typing method, and xj
is the number of isolates that belong to subtype xth.
The patterns that were found were compared
with those recorded in the international database
(http://mac-inmv.tours.inra.fr/) of MIRU-VNTR for
MAC and were assigned certain pattern number,
INMV22, 26, 27.
Results
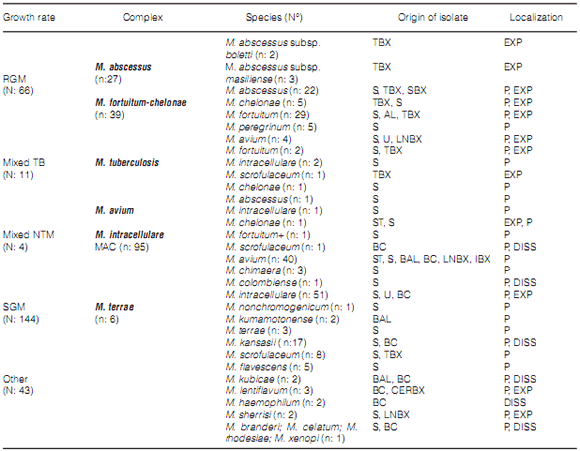
During the study period, the mycobacteriology laboratory of the Hospital Dr. Antonio A. Cetrángolo, as provincial model, diagnosed 4,236 cases of mycobacterial disease. 3,965 patients (93.9%) were confirmed as cases of TB: one caused by the bacillus BCG (Bacillus Calmette-Guérin), three by M. bovis and the rest, M. tuberculosis. In 271 cases (6.8%), one NTM was isolated as a potential causative agent of the disease. A total of 46 cases (16.9%) were excluded from the study: one NTM was isolated only one time in 31 cases (10 isolates belonged to the M. gordonae complex) and was discarded as causative agent of the disease27. We could not obtain the adequate identification of the isolate in 6 cases, and patient information was incomplete in 9 cases. 66 of the remaining 225 cases (29.3%) had RGM, and 159 (70.7%) had SGM. Two different mycobacteria species were isolated in 15 cases (6.7%) during the same disease episode. Figure 1 and Table 2, respectively, show the frequency of appearance of mycobacteriosis caused by SGM and RGM during the study years, and the differentiation of species and complexes of the isolates that were found. Table 3 shows the characteristics of the patients included in the study. 71.1% of the cases presented pulmonary localization, 38.8% of them with bacilli in the direct examination (AARB + [acid-alcohol resistant bacilli]); 8 patients (3.6%) presented disseminated disease with bacilli isolated from serial blood cultures; 6 of those patients were coinfected with HIV. 78 patients (34.7%) showed history of previously treated TB. 151 cases (67.1%), showed, both individually or simultaneously: a) HIV infection (43 patients); b) a particular physical condition (old age, pregnancy); c) one diagnosed comorbidity (110 cases, for example, diabetes, bronchiectases with and without cystic fibrosis, a malignant disease).
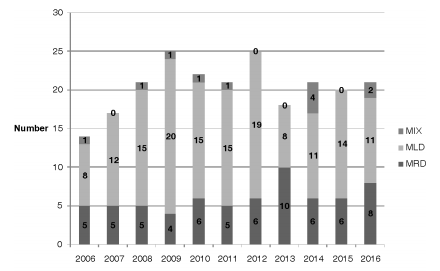
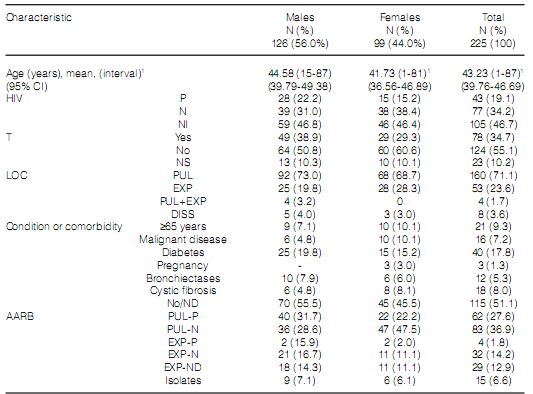
Despite the gender of the patient, we observed
a greater probability to develop mycobacteriosis
by M. intracellulare in patients with previous
treatment (odds ratio: 2.71487, 95% CI [confidence interval]: 1.087-6.798; X2: 4.398, p: 0.0324),
whereas the patients with HIV infection faced a
greater risk to develop the disease by M. avium subsp. hominissuis (odds ratio: 7.111, 95% CI:
1.542-33.17; X2: 7.930, p: 0.0125).
A total of 22 different species were recovered, 4
of which were RGM, and isolated from 66 patients
(29.3%): 27 cases (40.9%) with the M. abscessus complex, where 3 isolates belonged to the subspecies M. masiliense and 2 belonged to M. bolletii;
and 39 cases (59.1%) with the M. fortuitumchelonae complex, where 31 (47.0%) belonged to M. fortuitum, 4 belonged to M. chelonae and 4 to M. peregrinum.
The SGM included the remaining 18 species.
The MAC was isolated in 95 cases (42.2%), 40 of
them with M. avium subsp. hominissuis, 51 with M. intracellulare, 3 with M. Chimaera and 1 with M. colombiense.
Table 2 shows the number and frequency of
appearance of all the identified species, the cases attributed to mixed infection and the localization
of the disease that produced the isolates of the
different species.
11 of a total of 27 M. abscessus cases (40.7%)
were related to pulmonary disease, whereas the remaining 16 were obtained from skin lesions and/or
injuries caused by surgical or cosmetic procedures.
The M. avium subsp. hominissuis strains
presented pulmonary, extrapulmonary and disseminated localization. M. intracellulare did not
cause a disseminated disease in the study cases,
but cases of disseminated disease were proven with M. kansasii. The species M. flavescens, M. kubicae,
M. lentiflavum, M. haemophilum, M. sherrisi, M.
branderi, M. celatum, M. rhodesiae and M. xenopi were capable of producing pulmonary, extrapulmonary and disseminated disease according to
the origin of the samples from which the bacteria
were isolated and the clinical data of the patients.
A total of 15 cases presented mixed infection. In
11 patients, M. tuberculosis associated with other
species was isolated. In 4 HIV-positive cases, the
second germ that was found was M. avium subsp. hominissuis; in 2 HIV-positive cases, the second
germ was M. intracellulare and in HIV-negative
patients we found the following associations: 1
case with M. abscessus, 2 cases with M. fortuitum, 1 case with M. chelonae and 1 case with M. kubicae. The last case was an old patient at a nursing home.
In other 4 cases, microbiological findings were
produced generally during the treatment for the
first isolated germ. These 4 cases were:
a) Old patient: in 2010, M. fortuitum was isolated
and in 2011, before ending the corresponding
treatment, M. intracellulare was isolated.
b) HIV-positive patient: M. avium subsp. hominissuis was isolated and during the specific treatment
in the same year, M. intracellulare was isolated.
c) HIV-positive patient: M. chelonae and M. avium subsp. hominissuis were simultaneously isolated in the same disease episode.
d) HIV-positive patient: M. intracellulare was isolated from a sputum sample in 2008. In 2009,
during the treatment for mycobacteriosis, M.
scrofulaceum was isolated from a blood culture
sample.
In these 15 cases, mixed infection was associated with the history of previous and/or current
treatment in the male group (odds ratio: 1.5933
(95% CI: 0.6096-3.8217).
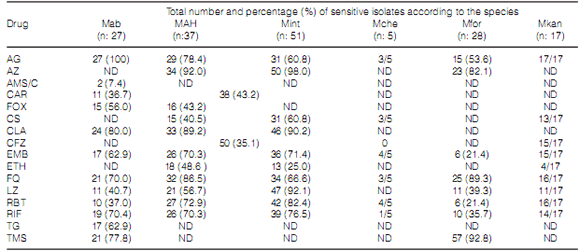
Table 4 shows the number and percentage of
isolates of every species sensitive to each one of
the drugs detailed in Table 1. EMB and RIF were
the only first line drugs used in TB treatment to
be included in the NTM antibiograms. RIF was
tested in a total of 165 isolates (75.7%). 109 cases
(66.0%) were sensitive to the drug. The EMB
showed 84.8% global inhibitory activity (140/165
isolates), especially toward M. kansasii and some
strains of MAC and M. chelonae. M. abscesus,
MAC and M. kansasii were mostly susceptible to
CLA, and M. foruitum and MAC to AZ. The CFZ
showed activity in over one third of the MAC
strains, whereas M. kansasii proved to be very
sensitive to this antibiotic. The FQs (levofloxacin and moxifloxacin) under evaluation inhibited more
than 70% of the species. From the beta-lactamic
antibiotics, AMS, AMC and FOX, only FOX was active against the M. abscessus complex. Two thirds
of these strains were inhibited by AG, FOX, CLA,
FQ, RIF and TMS, whereas a lower percentage
(around 60%) were also inhibited by EMB and TG.
Most of the MAC strains were inhibited by AG, AZ,
CLA, EMB, FQ, RIF and RBT. The activity of LZ
was higher on M. intracellulare isolates than on M. avium isolates.
80% of the MAH (n: 32) were properly genotyped, exhibiting great genetic diversity. With
the MIRU-VNTR technique, we found 5 known
patterns and other 6 patterns recently published
by our group, not previously reported in the literature28. X3 and 292 loci showed the highest discrimination index (D) among the MAH isolates (0.7424
and 0.6515), obtaining a high HGDI (0.9697).
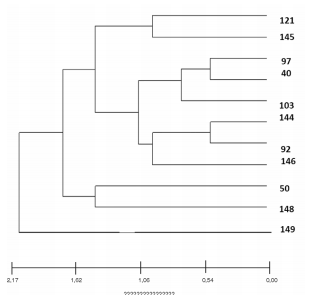
Figures 2 and 3 show the genetic patterns
(INMV) of the MAH that were found and the
existent phylogenetic relationship among them.
Relationships among the different INMV patterns
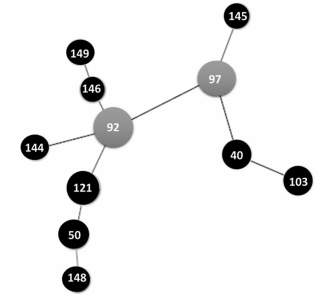
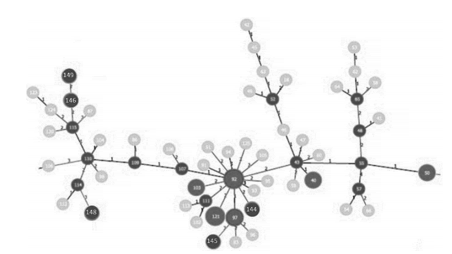
are shown in a dendrogram in Figure 2. Figures 3and 4 show clonal relationships among the different
patterns, represented by the minimum spanning
tree (MST) created from the goeBURST algorithm.
It is generally assumed that the genetic distance
between two INMV patterns is proportional to the
difference in the number of repetitions of each locus. These relationships were established using the Phyloviz 2 software (http://goeburst.phyloviz.net)26.
Regarding the success of the specific treatment,
we evaluated a total of 109 (63.7%) out of the 171
patients (76.0%) who began the treatment in the
2009-2014 period. In 71 cases (65.1%), the cohort
showed “ended treatment” or “healed” (ET/H).
Therapy suspension and deaths represented
12.8%, with 8 and 6 patients, respectively. There
were 6 relapses (5.5%) and 16 cases (14.7%) were
labeled as “continuing treatment” at the moment
of the evaluation. No information was obtained
about the remaining 62 patients (36.3%).
Of the 54 patients (24.0%) who initiated the
treatment in 2015 and 2016, it was possible to
evaluate treatment success in 41 patients (76.0%).
3 of them died (7.3%), 4 (9.8%) were registered as
ET/H and 34 (83.0%) were still under treatment
during the data collection phase. Of the 11 cases with mixed infection, 7 were registered as ET/H.
The treatment global success was estimated at
50.0% according to the relationship between the
number of cases evaluated in the 2009-2015 period
(n: 150) and the ET/H cases (n: 75).
Conclusions
NTM are ubiquitous, naturally free-living organisms, considered as opportunistic infectious agents, especially in hosts with cellular immunity disorders, where they produce a disease similar to TB,
generically called mycobacteriosis28.
The incidence of NTM was about 6.0% taking
into account all the cases initially considered as
TB, with a kind of stable tendency during the
period under evaluation. These results, in comparison with the ones previously published show
a stable tendency in the appearance of MB in our
environment20.
In this study, patients mostly presented lesions
with a respiratory localization, though the disease
can also be extrapulmonary, mixed or disseminated; over one third of patients had previously
received treatment for TB and almost half of the
patients presented comorbidities as base disease.
This information agrees with previously published
data29-31. However, previous TB was the most important risk factor for mycobacteriosis, especially
for the cases caused by M. intracellulare which,
together with M. avium subsp. hominissuis were
the most commonly found pathogens.
In total, 22 different species of NTM were recovered, where SGM predominate.
RGM, mainly the members of the M. abscessus complex, are considered emerging opportunistic
pathogens that may cause chronic pulmonary infections and diseases of the skin, soft tissues, bones
and joints. There are numerous publications where these RGM are involved in infections associated
with traumatic injuries, surgeries and various procedures: acupuncture, liposuction, mesotherapy,
and breast implant surgery29-31.
In this study, the number of cases with pulmonary localization of the disease caused by the M.
abscessus complex isolated from tissue and sputum
samples was similar to the number of cases with
extrapulmonary localization. M. nonchromogenicum and M. peregrinum were also found, but with
low frequency and with pulmonary localization
exclusively.
The species of the M. abscessus and M. fortuitum-M. chelonae complexes were the most frequently isolated among the RGM. The species of
the M. fortuitum-M. chelonae complex were mostly
related to pleuropulmonary diseases.
Within the MAC, the most commonly found
pathogens were M. intracellulare and M. avium subsp. hominissuis, though M. chimaera and M.
colombiense were also isolated. The members of the
MAC, together with M. kubicae and M. haemophilum caused the disseminated disease. M. avium subsp. hominissuis presented great genetic variability, suggesting a diversity of infection sources.
Also, previous TB was the most important risk
factor, especially for the cases where the disease
had been caused by M. intracellulare.
Other species of SGM such as M. kumamotonense and M. rhodesiae were also found, though
in small proportion.
Some NTM, such as the members of the MAC,
are able to infect not only birds but also mammals,
for example, pigs. In a study recently published in
Belgium, more than 98% of pig isolates belonged
to M. avium subsp. hominissuis. This finding
could suggest the zoonotic transmission of these
mycobacteria25. This makes us think about the
need to plan genetic and epidemiologic studies
to corroborate or discard potential zoonosis in
mycobacteria and also to evaluate possible direct
transmission between hosts.
Although the results of NTM antibiograms
are informative and must be carefully evaluated
by attending physicians, the results of this study
showed concordance with the information previously published by several authors31-33. In general
terms, of all the drugs under evaluation, aminoglycosides, fluoroquinolones and macrolides were
the most active against a greatest variety of NMT, but there wasn´t any species homogenously sensitive to said drugs. The other drugs showed a more
evident species-dependent relationship. In this
regard, the identification of the germ at the species
level is an important predictor of its sensitivity
to drugs and a guide to aid the physician in the
implementation of a suitable therapy. A review has
been recently published about new antimicrobials
active against NTM at several growth phases. In
this study, the evaluation of cohorts showed that
around half of the patients with mycobacteriosis
healed, though an important loss of information
was registered and the cohorts were incomplete.
A remarkable fact was to find that both M.
tuberculosis and NTM were causing the disease.
The simultaneous isolation of M. tuberculosis together with NTM was considered as cause of joint
disease. The patients were treated with regularized regimens for TB using, in the early phase,
isoniazid, rifampicin, etambutol and pyrazimide,
to which M. tuberculosis isolates were originally
sensitive. They continued with conventional treatment until detection of NTM and/or treatment
failure (positive smear tests and cultures) resulted
in the customization of therapeutic regimens, by
adding aminoglycosides, fluoroquinolones and
any other drug depending on the identified NTM
species (Tártara, personal communication), the
recommended regimens and the in vitro sensitivity
results obtained from the isolates33. Most of these
patients healed.
Regarding the variability of the M. avium subsp. hominissuis isolates obtained, the molecular technique we used allowed us to show the existence of
great genetic diversity among the isolates. Also,
the INMV patterns obtained in this study could
be compared and analyzed with the ones reported
by other authors in the international database
(MAC-INMV)24. The creation of the MST allowed
us to establish clonal relationships among the different isolates. INMV 92 turned out to be a native
clone, both when the MST included only the study
isolates and also when it included the isolates globally reported in the database. This confirms its
role as a clone from which other M. avium subsp.hominissuis isolates derive.
Acknowledgement. Marcelo Mazza, Guillermo Alonso, Bibiana Zingoni and Claudia Palavecino for their excellent technical work.
Conflict of interest: The authors of this work declare there is no conflict of interest related to this publication. BRI, MPS and MIR are researchers at CONICET; DM is a postdoctoral fellow at CONICET.
1. Nunes-Costa D, Alarico S, Pretti Dalcolmo M, Correia-Neves M, Empadinhas N. The looming tide of nontuberculous mycobacterial infections in Portugal and Brazil. Tuberculosis 2016; 96: 107-119.
2. Stahl, DA, Urbance JW. The division between fast- and slow-growing species corresponds to natural relation-ships among the mycobacteria. J Bacteriol 1990; 172: 116-124.
3. Tortoli E. Microbiological features and clinical relevance of new species of the genus Mycobacterium. ClinMicro-biolRev 2014; 27: 727e52. En: http://dx.doi.org/10.1128/CMR.00035-14.
4. Brown-Elliot BA, Griffith De, Wallace RJ. New described emerging human species of nontuberculous mycobacteria. Infect Dis Clin N Am 2002; 16:187-220.
5. Shinnick TM, Good RC. Mycobacterial taxonomy. Eur J Clin Microbiol Infect Dis 1994; 13: 884-901.
6. Tsang AY, Barr VL, McClatchy JK, Goldberg M, Drupa I, Brennan PJ. Antigenic relationships of the Mycobacterium fortuitum- M. chelonae complex. Int J Syst Bacteriol 1984. 34: 35-44.
7. Brown-Elliott BA, Wallace RJ Jr ., Tichindelean Cet al. Five-Year Outbreak of Community and Hospital-Acquired Mycobacterium porcinum Infections Related to Public Water Supplies. J Clin Microbiol 2011; 49: 4231-4238.
8. Sassi M, Drancourt M. Genome analysis reveals three genomospecies in Mycobacterium abscessus. BMC Genom- ics 2014; 15:359 En: http://www.biomedcentral.com/14712164/15/359.
9. Tortoli E, Kohl TA, Brown-Elliott BA et al.Emended description of Mycobacterium abscessus, Mycobacterium abscessus subsp. abscessus and Mycobacterium abscessus subsp. bolletii and designation of Mycobacterium abscessus subsp. massiliense comb. Nov. International Journal of Systematic and Evolutionary Microbiology. 2016; 66: 4471–4479.
10. Tan JL, Ngeow YF , Choo SW. Support from Phylogenomic Networks and Subspecies Signatures for Separation of Mycobacterium massiliense from Mycobacterium bolletii. J Clin Microbiol 2015; 53: 3042-3046.
11. Brown-Elliott B A, Wallace RJ. Clinical and Taxonomic Status of Pathogenic Nonpigmented or Late-Pigmenting Rapidly Growing Mycobacteria. Clinical Microbiology Reviews2002; 15: 716–746.
12. Narnaware SD, Periasamy S, Tripathi BN. Studies on pathology, cytokine gene expression and molecular typing of Mycobacterium avium subsp. paratuberculosis of naturally occurring Johne’s disease in bullocks. Res Vet Sci. 2016; 106:74-80.
13. Kan P, Pilli AS, Rampal R et al. Prevalence and Association of Mycobacterium avium subspecies paratuberculosis with disease course in patients with ulcero-constrictive Ileocolonic disease.PLoS One. 2016; 11(3): e0152063.doi: 10.1371/journal.pone.0152063.
14. Griffith DE, Aksamit T, Brown-Elliot BA, Cattanzaro A, Daley C, Gordon F. An official ATS/IDSA statement diagnosis, treatment and prevention of nontuberculous mycobacterial disease. Am J Respir Crit Care Med 2007; 175: 367-416.
15. American Thoracic Society. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. Am J Respir Crit Care Med 1997; 156:S1–S25.
16. Tortoli E. The impact of whole genome sequencing on the taxonomy of mycobacteria. European Society of Mycobacteria, 37th Annual Congress, 3rd to 6th July 2016, Catania, Sicily, Italia. Scientific Program and Abstracts Book, pp: 29.
17. Becton Dickinson. BBL MGIT Products for the Detection of Mycobacteria. Becton Dickinson Cockeysville, Maryland, USA. Insert 88-0950, 1996.
18. Jacomo V, Musso D, Gevaudan MJ. Isolation of blood-borne Mycobacterium avium by using the nonradiactive BACTEC 9000 MB system and comparison with a solid-culture system. J ClinMicrobiol. 1998; 3703-3706.
19. Martin A, Paasch F, Docx S, et al. Multicentre laboratory validation of the colorimetric redox indicator (CRI) assay for the rapid detection of extensively drug-resistant (XDR) Mycobacterium tuberculosis. J Antimicrob Chemother 2011; 66: 827-833.
20. Richter E, Rüsch-Gerdes S, Hillemann D. Evaluation of the GenoType Mycobacterium Assay for Identification of Mycobacterial Species from Cultures. J Clin Microbiol. 2006; 44: 1769-1775.
21. Imperiale B, Zumárraga M, Gioffré A, Di Giulio B, Cataldi A, Morcillo N. Disease caused by non-tuberculous mycobacteria: diagnostic procedures and treatment evaluation in the North of Buenos Aires Province..Rev Argent Microbiol2012; 44:3-9.
22. Travería G, Zumárraga M, Etchechoury I et al. First identification of Mycobacterium avium paratuberculosis sheep strain in Argentina. Brazilian J Microbiology 2013; 44: 897-899.
23. Thibault VC, Grayon M, Boschiroli ML et al. New variablenumber tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing.J Clin Microbiol 2007; 45: 2404-2410.
24. Imperiale B, Moyano RD, Di Giulio AB et al. Genetic diversity of Mycobacterium avium complex strains isolated in Argentina by MIRU-VNTR. Epidemiology and Infection, Cambridge University Press 2017, pp: 1-10 doi: 10.1017/S0950268817000139.
25. Vluggen C, Soetaert K, Duytschaever L et al. Genotyping and strain distribution of Mycobacterium avium subspecies hominissuis isolated from humans and pigs in Belgium, 2011-2013. Euro Surveill. 2016; 21: 30111. doi: 10.2807/1560-7917.ES.2016.21.3.30111.
26. Hunter P. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol 1990; 28: 1903-1905.
27. Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J ClinMicrobiol1988; 26:2465- 2466.
28. Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramírez M, Carric JA. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 2012, 13:87.DOI: 10.1186/1471-2105-13-87.
29. Wassilew N, Hoffmann H, Andrejak C, Lange C. Pulmonary Disease Caused by Non-Tuberculous Mycobacteria. Respiration 2016;91:386–402. DOI: 10.1159/000445906.
30. Rivera-Olivero IA, Guevara A, Escalona A et al. Infecciones en tejido blando debidas a micobacterias no tuberculosas posterior a mesoterapia. ¿Cuál es el precio de la belleza? Enferm Infecc Microbiol Clin. 2006; 24: 302-306.
31. Da Mata O, Hernández-Pérez R, Corrales H, Cardoso-Leao S, de Waard J. Seguimiento de un brote de infección en tejido blando causado por Mycobacterium abscessus posterior a la mesoterapia en Venezuela. Enferm Infecc Microbiol Clin 2010; 28:596-601.
32. I. Porvaznik, I. Solovicˇ J, Mokry J. Non-Tuberculous Mycobacteria: Classification, Diagnostics, and Therapy. Advs Exp. Medicine, Biology. Neuroscience and Respiration 2016. DOI 10.1007/5584_2016_45.
33. Soni I, De Groote MA, Dasgupta A, Chopra S. Challenges facing the drug discovery pipeline for non-tuberculous mycobacteria. J Medical Microbiol2016; 65: 1–8.
34. Brown-Elliott BA, Nash KA, Wallace RJ Jr. Antimicrobial Susceptibility Testing, Drug Resistance Mechanisms, and Therapy of Infections with Nontuberculous Mycobacteria. Clinical Microbiology Reviews. CMR J Association for Microbiology 2012; 25: 545-582.